Ethical statement
This study was approved by the Ethics Committee of West China Hospital, Sichuan University (Approval Number: 2020-1151). All participants provided informed consent for the sharing of their transcriptomic data and were made aware of any associated risks. The open access use of these data was permitted by the Ministry of Science and Technology of China (Registration Number: 2022BAT2228).
Participant information
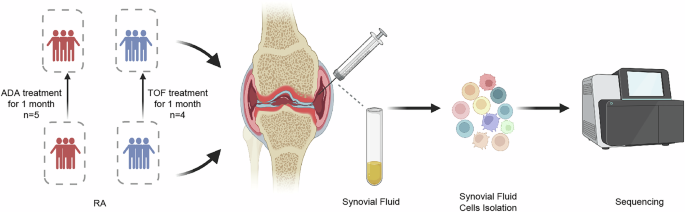
In our study, nine patients who met the 2010 revised criteria14 for RA established by the American College of Rheumatology were prospectively and randomly enrolled from department of rheumatology and immunology, West China Hospital, Sichuan University. Informed consent was obtained from all participants. At baseline, all patients were required to exhibited high or moderate disease activity and to have an inadequate response to methotrexate. In addition, these patients were required not received glucocorticoids or any bDMARDs or tsDMARDs within 3 months prior to enrollment. Clinical and demographic details are provided in Supplementary Table 1-2. To comprehensively define the transcriptional atlas of SF cells from RA patients, who are MTX insufficient responders, we performed scRNA-seq on nine paired samples from RA patients before and after one month of treatment with TNF-α or JAK inhibitors (referred to as RA-BT and RA-AT, respectively). Specifically, nine enrolled RA patients were treated with either adalimumab (n = 5, 40 mg every 2 weeks) or tofacitinib (n = 4, 5 mg twice a day) in conjunction with a stable dose of MTX. A second knee arthrocentesis was performed one month later to collect SF cells from the same joint.
SF cells isolation
The patient’s knee joint synovial fluid was extracted under aseptic conditions. Hyaluronidase (20426ES60, Yeasen) was added to the fresh SF sample obtained at a final concentration of 100 U/ml and incubated at 37 °C for 30 min. SF samples from each RA patient were obtained from the same joint longitudinal to avoid potential bias.
Cell suspension preparation
To prepare single-cell suspensions for sequencing needs, the fresh SF samples were centrifuged at 800 × g for 10 min, the supernatant was collected and stored at −80 °C, while the cell pellets were suspended by 1 ml red blood cell lysis buffer (R1010, Solarbio) to lyse the red blood cells and incubated on ice for 6 min, followed by centrifugation at 800 × g for 5 min. Next, 800 μl of pre-chilled PBS (BL302A, Biosharp) and 300 μl of pre-chilled debris removal solution (130-109-398, Miltenyi; reagent for cell debris removal through density gradient centrifugation) were added to suspend the cell pellets. After gently adding additional 800ul PBS and centrifuging at 800 × g for 5 min, the liquid will be divided into three phases. The top two phases were discarded, and 2 ml PBS were added to wash the lower phase of pellets once, centrifugation was performed at 800 × g for 5 min to remove contaminants and obtain pure cells. Finally, we collected a 20 μl aliquot for cell counting after discarding the supernatant and resuspended it by adding 500 μl PBS. All centrifugation procedures were carried out at 4 °C. Finally, high-quality single-cell suspensions were prepared for scRNA-seq.
Single cell library preparation and sequencing
The cell suspension was placed into chromium microfluidic chips, and a 10 × Chromium Controller (10× Genomics) was used to barcode the samples. Next, using components from a Chromium Single Cell 3’ (v3) reagent kit (10× Genomics), sequencing libraries were created using RNA from the barcoded cells after reverse transcription, in accordance with the manufacturer’s instructions. Sequencing was carried out using the Illumina NovaSeq 6000 platform in accordance with the manufacturer’s instructions. Single cell library preparation and subsequent next generation sequencing was performed by Novogene Co., Ltd. All the scRNA-seq data has passed the quality control.
Data processing and analysis
The scRNA-seq data was analyzed using our previously established workflow15,16,17,18,19,20,21,22,23. Briefly, raw FASTQ files generated by 10× Genomics were aligned and quantified using Cell Ranger (v4.3.0) against the GRCh38 human reference genome (v6.1.2). The Read10X function of the Seurat package was used to read the output of Cell Ranger. Doublets were identified and removed using the scrublet tool in Python, with an expected doublet rate of 0.1. Next, to facilitate downstream analyses, cells were distinctly labeled using the RenameCells function and integrated into a single aggregate object using the Merge function. To ensure data quality, stringent filtering parameters were applied to weed out any empty oil beads, mortality, and doublet cells, excluding cells with fewer than 200 or more than 4,000 detected genes, or with mitochondrial content exceeding 15%. Global scaling normalization was performed using the ‘LogNormalize’ method with a scaling factor of 10,000 to equalize overall gene expression across cells. The FindVariableFeatures function identified the top 2,000 highly variable genes, which were used for dimensionality reduction via principal component analysis (PCA). The first 30 principal components were selected, and the ScaleData function was applied to scale the expression of all genes, followed by regression of mitochondrial content to remove confounding variation. Sample batch effect correction was performed using the RunFastMNN function24, which is based on a multi-canonical correlation analysis algorithm. For clustering, the FindNeighbors and FindClusters functions with the built-in Louvain method were applied. Clusters were visualized using uniform manifold approximation and projection (UMAP). Cluster-specific marker genes were identified using the non-parametric Wilcoxon rank-sum test with Bonferroni correction (adjusted p value < 0.05) implemented in the FindAllMarkers function (min.pct = 0.25, logfc.threshold = 0.25). Cell populations were annotated based on canonical marker genes from published literature. Due to the inherent instability of neutrophils during processing and their predominant origin from a small number of samples, this cell type was excluded from subsequent analyses to mitigate technical bias. The proportion of each cluster was estimated by dividing the count of cells within each cluster by the total cell count in the respective sample. These ratios were statistically compared across treatment groups to assess significant differences in cell composition. The detailed workflow for data processing, including the steps for transferring data and generating figures and analyses presented in this study, is available on our GitHub repository at (https://github.com/Xiaxy-XuLab/RA-SF-data_discriptor).