Study approval
This study does not involve sex-based analysis, and sex was not considered in the study design, since RA involves both. This study was approved by the Ethics Committee of Wenzhou Medical University. Informed consent was obtained from patients before inclusion in the study. The human studies were adhered to the Declaration of Helsinki. Animal experiment procedures were approved by the Institutional Animal Care and Use Committee of Wenzhou Medical University. All animal experiments in this study abide by the ARRIVE guidelines.
Patients and samples
Serum and synovial fluid samples were obtained from active RA patients who were newly diagnosed in the outpatient department before disease-modifying antirheumatic drug (DMARDs) treatment, serum from RA patients in remission who were treated with DMARDs. And synovial tissues were obtained from RA patients underwent knee arthroplasty who have not been given biological agents. Seropositive refers to RA patients who test positive for RF or ACPA, and seronegative refers to RA patients who test negative for both RF and ACPA. All RA patients fulfilled the 2010 American College of Rheumatology/European League Against Rheumatism (ACR/EULAR) criteria. Synovial tissues were obtained from GA patients fulfilled the 2015 ACR/EULAR gout classification criteria. Serum obtained from PsA patients fulfilled the Classification Criteria for Psoriatic Arthritis. Serum of HC from healthy volunteers, and the synovial tissues, synovial fluid and cartilage were used to be controls from patients of non-inflammatory knee joint diseases underwent knee operations. FLSs were isolated from synovial tissues as previously described54. In brief, after obtaining the synovial tissue, large pieces of adipose tissues were removed, the blood was washed off, and in a sterile environment, the synovial tissue was repeatedly cut into a paste-like consistency and centrifuged to remove adipose tissues. Finally, FLS were isolated through enzymatic digestion using type II collagenase. All FLS from passage 3 to 5 were used for the experiments. This study was approved by the Ethics Committee of Wenzhou Medical University. Informed consent was obtained from patients before inclusion in the study. The clinical details of all participants are shown in Supplementary Table S1.
Cells culture, intervention and transfection
FLSs from HC, GA patients and RA patients were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, USA) supplemented with 10% Fetal Bovine Serum (FBS, 11011-8611, Ever Green, China) at 37 °C in 5% CO2.
In the intervention experiments, RA-FLSs were treated with 10 mM lactate (L1750, Sigma-Aldrich, Germany), 3% oxygen, 10 ng/mL human TNF-α (10291-TA, R&D Systems, USA), 1 ng/mL human IL-1β (201-LB, R&D Systems, USA), 10 mM sodium lactate (L7022, Sigma-Aldrich, Germany), and hydrochloric acid, respectively. After 24 h incubation, the cells were collected for western blotting. In the inhibitor intervention experiments, RA-FLSs were pre-treated with 10 µM lactate dehydrogenase inhibitor (FX-11, 213971-34-7, MCE, USA) or 10 mM lactate+100 nM MCT1 inhibitor (AZD-3965, 1448671-31-5, MCE, USA) for 24 h, and the cells were used for western blotting, ChIP-qPCR, RT-qPCR, scratch migration and transwell assay. Cell viability of FLSs were detected using Calcein/PI Cell Viability/Cytotoxicity Assay Kit (C2015M, Beyotime, China) according to the manufacturer’s instructions. Finally, images of the live (green fluorescence) and dead (red fluorescence) cells were captured using an inverted fluorescence microscope (Ti-S, Nikon, Japan).
In the siRNA transfection experiment, the cells were cultured into 6-well plates (2.5 × 105 cells/well) for 24 h. Then, siRNA diluted with 250 µL Opti-MEM medium (11058021, Gibco, USA) was mixed with lipofectamine 3000 (L3000015, Thermo Fisher Scientific, USA) diluted with 250 µL DMEM. Then, the mixture was added to each well. After 6 h culture, the supernatant was replaced by basal medium and cultured for additional 48 h. The cells were collected for western blotting, ChIP-qPCR, RT-qPCR, scratch migration and transwell invasion assay. The siRNA sequence was described in Supplementary Table S3.
Western blotting
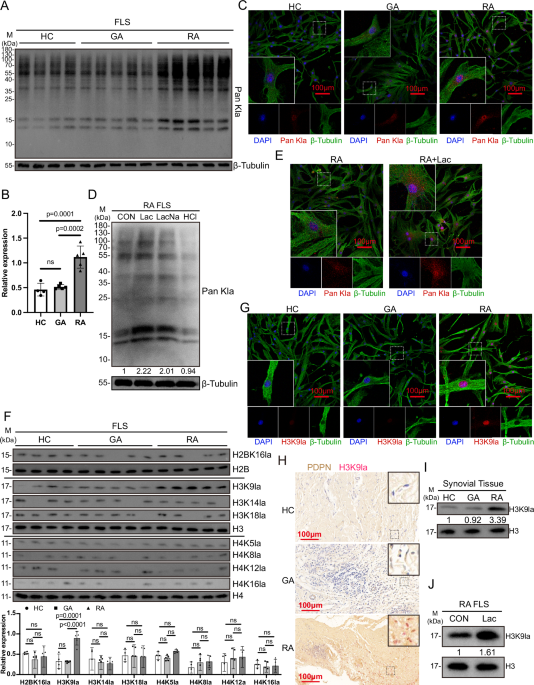
FLSs or synovial tissues were lysed with RIPA lysis buffer. The protein was quantitated by BCA Protein Assay Kit (P0012, Beyotime, Shanghai, China). 20 μg protein were loaded onto Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels for separation and then transferred to polyvinylidene difluoride (PVDF) membranes. The membranes were blocked and incubated with the primary antibodies at 4 °C overnight, and then incubated with the HRP-conjugated secondary antibodies. The bands were detected by electrochemiluminescence (ECL), and the ChemiScope 6000 imaging system (Clinx, China) was applied for photography. Protein gray value detection was analyzed by Image J software. The following antibodies used were available from Supplementary Table S4.
Immunofluorescence staining and immunohistochemistry
FLSs were inoculated on the round coverslip and fixed with 4% paraformaldehyde after adhesion, then were blocked with 5% BSA and incubated with primary antibody overnight at 4 °C. On the second day, incubated with fluorescein-conjugated secondary antibody for 1 h in the dark at room temperature, nuclei were counterstained with DAPI.
The mouse joint sections or synovial tissue sections were deparaffinized, hydrated, antigenically repaired, blocked, and incubated with primary antibody overnight at 4 °C. For immunofluorescence analysis, then incubated with fluorescein-conjugated secondary antibody for 1 h in the dark at room temperature, nuclei were counterstained with DAPI. Imaging with laser scanning confocus (C2si, Nikon, Japan). For immunohistochemistry analysis, incubated with HRP-conjugated secondary antibody after incubated with primary antibody, then visualization with the 3,3′-Diaminobenzidine (DAB) substrate and imaging with photomicroscope (CI-L, Nikon, Japan). The following antibodies used were available from Supplementary Table S4.
Lactate measurement
The concentration of lactate in synovial fluid was detected using L-Lactate Assay Kit (ab65330, Abcam, USA) according to the manufacturer’s instructions. The quantification was performed with Varioskan LUX multimode microplate reader (Thermo Fisher Scientific, USA) at 570 nm.
ELISA
The concentrations of TNF-α, IL-1β in synovial fluid and the concentrations of IL-1β, TNF-α, IL-6, IL-10 in the serum of mice were detected using ELISA kits (R&D Systems, USA). The concentrations of IL-6, IL-8, MMP9 and MMP13 in supernatant from RA-FLS were detected using ELISA kits (Abcam, UK). As per the manufacturer’s instructions and measured using Varioskan LUX multimode microplate reader at 450 nm.
To detect anti-lactylated histone in serum, the Costar ELISA plates (Corning, USA) were coated with peptides (5 µg/mL) in PBS at 4 °C overnight. Subsequently blocked by 2% BSA (1 h, room temperature) and following incubation with human serum (diluted 1:100) for 1 h at room temperature. After three times washed with PBST, followed by HRP-Goat anti-human IgG (AS002, ABclonal, Wuhan, China) for 1 h at room temperature. Bound antibodies were visualized using tetramethylbenzidine and hydrogen peroxide. Measured using Varioskan LUX multimode microplate reader at 405 nm.
CUT&Tag assay and bioinformatics analysis
To explore the candidate target genes of H3K9la in FLSs, CUT&Tag assay was conducted using Hieff NGS G-Type In-Situ DNA Binding Profiling Library Prep Kit for Illumina (12598, Yeasen, China), following the manufacturer’s instruction (three independent samples of each group). Briefly, cells were harvested and bound to concanavalin A-coated magnetic beads, reacted with primary antibody against H3K9la, secondary antibody of anti-rabbit IgG, and pA/G-Transposome Mix to activate transposase. FLSs were lysed by proteinase K and DNA was extracted, amplified and purified for library construction using Hieff NGS Tagment Index Kit (12416, Yeasen, China), and the library quality was assessed on the Agilent Bioanalyzer 2100 system. Then the library was sequenced on Illumina Novaseq platform at Novogene Science and Technology Co., Ltd (Beijing, China). The peaks with fold change of RPM more than 2, p < 0.05 were considered as differential peaks. The position of peak summit around transcript start sites of genes can predict the interaction sites between protein and gene. ChIPseeker55 was used to retrieve the nearest genes around the peak and annotate genomic region of the peak. Peak-related genes can be confirmed by ChIPseeker, and subjected to enrichment analysis of KEGG. Motif analysis was performed using findMotifsGenome.pl program in HOMER v4.11 software.
RNA-seq and bioinformatics analysis
The total RNA of FLSs (three independent samples of each group) was extracted using Trizol reagent (15596026, Invitrogen, USA) following the manufacturer’s procedure. The RNA libraries were sequenced on the Illumina NovaseqTM 6000 platform by LC Bio-Technology CO.,Ltd (Hangzhou, China). Differentially expressed genes (DEGs) were screened by DESeq2 software between RA-FLS and HC-FLS. The genes with the parameter of q value < 0.05 and |log2FC|> = 1 were considered DEGs. Then DEGs were subjected to enrichment analysis of GO and KEGG. The critical value of significant gene enrichment was set as p < 0.05.
Chromatin immunoprecipitation (ChIP)-qPCR
The assay was performed using ChIP Kit (ab500, Abcam, USA) following the standard protocol. Briefly, FLS were fixed with 1% formaldehyde and quenched with 125 mM glycine, then the cells were lysed and sonicated. For immunoprecipitation, the diluted chromatin was incubated with H3K9la antibodies overnight at 4 °C with constant rotation. The precipitated chromatin DNA was purified with the QIAquick PCR Purification Kit (Qiagen). The qPCR was performed using Taq Pro Universal SYBR qPCR Master Mix (Q712, Vazyme, China) and analyzed using the QuantStudio 6 Flex Real-Time PCR system (Thermo Fisher Scientific, USA). The primer sequences were described in Supplementary Table S5.
RT-qPCR
Total RNA was isolated from FLSs using Trizol reagent (15596026, Invitrogen, USA). Reverse transcription was performed using HiScript III RT SuperMix for qPCR (R323, Vazyme, China). Real-time quantitative PCR (qPCR) was performed in QuantStudio 6 Flex Real-Time PCR system using Taq Pro Universal SYBR qPCR Master Mix. Primer sequences of NFATc2 and β-tubulin genes were described in Supplementary Table S4. The relative mRNA expression normalized to β-tubulin was calculated using the 2−ΔΔCt method.
Construction of short hairpin RNA-expression lentivirus and infection
The lentivirus-based shRNA vectors were designed and constructed by Tsingke Biotech Co., Ltd. (Beijing, China). The lentivirus of shRNA targeting NFATc2 and the control lentivirus were generated for the NFATc2-knockdown experiment. Briefly, pLKO.1 was constructed as the lentivirus transfer vector, which contained a puromycin resistant gene. The co-transfection of HEK 293T cells with shRNA-pLKO.1 and helper plasmids, psPAX2, and pMD2.G vectors. During transfection, transfected cells were screened using puromycin, and lentiviral particles were collected. For infection, RA-FLS were treated with lentiviruses and polybrene (10 µg/mL) for 12 h at 37 °C. Subsequently, the virus-containing medium was replaced with fresh medium, the cells were collected for the follower experiments. The transfection and infection efficiency were observed under an inverted fluorescent microscope, and the knockdown efficiency of shRNA was evaluated by WB. The sequences of shRNA were described in Supplementary Table S3.
Scratch migration assay
RA-FLSs were seeded in six-well plates (1 × 105 cells/well). the scratch was made along the diameter of the well using a 10 μL pipette tip (Axygen, USA) as the cells reached to 95% confluency. Images were captured at 0 h and 24 h by a camera under the microscope (×40 magnification). Image J was used to calculate the mobility ratio, and the mobility ratio was calculated using the following equation: migrated cellular area/scratched area × 100%.
Transwell assay
A 24-well transwell chamber with 8-μm pores (Corning, USA) was used for Transwell assays. The bottoms of the transwells (on top of the membrane) are coated with a thin layer of Matrigel (356234, Corning, USA). FLSs were trypsinized and re-suspended with serum-free DMEM medium at a final concentration of 1 × 105/mL, 200 μL cell suspension was seeded at the top of matrigel, the cells are cultured in the upper chamber of the transwell insert. 500 μL DMEM with 10% FBS as chemoattractant was added to the lower chamber. The plate was incubated at 37 °C under 5% CO2 for 24 h. Then, the migrated cells on the underside of the membrane were fixed with methanol, and stained with crystal violet. Pictures were taken under the microscope (TS100-F, Nikon, Japan) and the cell number was quantified by the software Image-Pro.
Humanized synovitis in vivo model using SCID mice
The cartilage from HC was cut into 5–8 mm3 pieces (one piece) and implanted into the left flanks of SCID mice wrapped with a sterile sponge infiltrated with RA-FLS (5 × 105 cells per 100 μL). In the right flanks, the cartilage (one piece) was implanted only. In order to observe the migration of FLSs on the left flanks to the cartilage on the right flanks, the mice were sacrificed on day 60 after implantation, the right implants were removed and embedded in Tissue-Tek OCT compound (4583, Sakura Finetek, Japan) for frozen section and hematoxylin-eosin (H&E) staining. Then the invasiveness of RA-FLS into cartilage was observed. The level of invasiveness was scored as follows: 0 = no or minimal invasion, 1 = visible invasion (two-cell depth), 2 = invasion (five-cell depth), and 3 = deep invasion (more than ten-cell depth).
Construction of CAIA model in NFATc2fl/fl × Thy1-CreERT2−/− and NFATc2fl/fl × Thy1-CreERT2+/− mice
All mice were raised in a specific-pathogen-free (SPF) room at the Laboratory Animal Center of WMU and housed in cages (five per cage) kept at 22–26 °C and 60–65% humidity on a regular 12-h light/dark cycle (light, 8:30–20:30) and had ad libitum access to water and food, except where noted. Rodent chow diet (corn, soybean meal, flour, bran, fish meal, salt, various vitamins and mineral elements, etc) were purchased from Keao Xieli Feed Co (Beijing, China). This study does not involve sex-based analysis, and sex was not considered in the study design. Therefore, in order to control for the variable, we used only male mice in this study. NFATc2fl/fl × Thy1-CreERT2−/− and NFATc2fl/fl × Thy1-CreERT2+/− mice (5 weeks old) were treated with tamoxifen (10540-29-1, Sigma-Aldrich, Germany) (Administer 175 mg/kg tamoxifen in corn oil via intraperitoneal injection once every 48 h for 3 times). Mice were tail intravenously day 18 with 5 mg anti-type II collagen monoclonal antibody cocktail (Chondrex, USA), according to the manufacturer’s instructions. The mean clinical score (0–4) was assigned as follows: 0 = no symptoms, 1 = erythema and slight swelling limited to the ankle joint and toes, 2 = erythema and slight swelling spreading from the ankle to the midfoot, 3 = erythema and severe swelling spreading from the ankle to the metatarsal joints, and 4 = ankylosing deformity with joint swelling. On day 25, The mice were euthanized, and collect serum and joint tissues for further study.
CIA model and histological analysis
DBA/1 mice were obtained from Shanghai SLAC Laboratory Animal. All experiment procedures were approved by the Institutional Animal Care and Use Committee of Wenzhou Medical University. All mice were raised in a SPF room at the Laboratory Animal Center of WMU and housed in cages (five per cage) kept at 22–26 °C and 60–65% humidity on a regular 12 h light/dark cycle (light, 8:30–20:30) and had ad libitum access to water and food, except where noted. Rodent chow diet (corn, soybean meal, flour, bran, fish meal, salt, various vitamins and mineral elements, etc) were purchased from Keao Xieli Feed Co (Beijing, China). This study does not involve sex-based analysis, and sex was not considered in the study design. Therefore, in order to control for the variable, we used only male mice in this study. On day 0, the mice were intradermally injected with 100 µL of type II bovine collagen (2 mg/mL) (20021, Chondrex, USA) emulsified in equal volumes of Freund’s complete adjuvant (7001, Chondrex). On day 21, the mice were injected with 100 µL of type II bovine collagen (2 mg/mL) emulsified in equal volumes of Freund’s incomplete adjuvant (7002, Chondrex) for booster immunization. Meanwhile, mice were given weekly intra-articular injections of shRNA (2 × 108 TU/mL) or daily intraperitoneally with FX-11 (2 mg/kg) or AZD-3965 (100 mg/kg). On day 49, the mice were euthanized, joint tissues and serum samples were harvested for analysis. The knee joints of mice were dissected and fixed in 4% paraformaldehyde and then decalcified in 50 nM EDTA and embedded in paraffin. The sections were deparaffinized, rehydrated, and stained with hematoxylin and eosin (H&E) or safranine O-fast green stain for histological analysis. Histology scores were assessed as previously described56.
Peptides synthesis
The peptides of lactylated histone and non-lactylated histone (10–20 aa) corresponding to the whole sequence of the human histone H2B, H3 and H4 were synthesized by KS-V Peptide Biotechnology Co., Ltd (Hefei, China). Peptides were purified by high-performance liquid chromatography (HPLC) (purity >95%) and characterized by electrospray ionization mass spectrometry (ESI-MS). The sequences of peptides were described in Supplementary Table S6.
Dot blot assay
The peptides were diluted into 1 µg/mL, then spotted (1 µL/dot) severally onto the nitrocellulose membrane (1 cm × 1 cm) using narrow-mouth pipette tip, then dried naturally and blocked by 10% BSA (2 h, room temperature), three times washed with PBST. Subsequently, the membrane was incubated overnight with human serum (200 µL) at 4 °C, three times washed with PBST, followed by HRP-Goat anti-human IgG (AS002, ABclonal, Wuhan, China) for 1 h at room temperature. The dots were detected by electrochemiluminescence (ECL), and the ChemiScope 6000 imaging system (Clinx, China) was applied for photography. Exposure time is controlled at 10 seconds.
Statistical analysis
Statistical analysis was conducted in GraphPad Prism 8 (GraphPad Software, La Jolla, CA), and the data were presented as the mean ± SD. The Shapiro–Wilk method was used to determine whether the data were normally distributed and the homogeneity of variance was tested by the Levene method. If the measurements between two groups were normally distributed, the unpaired Student’s t test was used, otherwise the Mann–Whitney U test was used. One-way analysis of variance test with post hoc contrasts by Tukey test was applied to compare the means of multi groups. ANOVA of repeated measurements was used for inference the effects of treatment and time on experimental objects (clinical scores of mice).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.