Study design
This study was approved by the clinical ethics committees of Hiroshima University Hospital, Dohgo Spa Hospital, Ehime University Proteo-Science Center, and Graduate School of Medicine (approval number: E-668; approval date: 01/02/2017). All experiments were performed according to the approved guidelines. Synovial tissues were collected from patients with both OA and RA who fulfilled the classification criteria of the 1987 American College of Rheumatology39 and underwent total joint replacement after providing informed consent.
Primary cultured and immortalized FLS preparation and culture
Primary cultured human FLS were isolated from the synovial tissue of patients with RA. Briefly, synovial tissues from total joint replacement lesions were minced and incubated with 1 mg/mL of collagenase/dispase (Roche, Mannheim, Germany) in phosphate-buffered saline (PBS) at 37 °C for 1 h, and subsequently filtered. For FLS preparation, the synovial cells were diluted and cultured. During culture, the supernatant was replaced frequently to dilute non-adherent cells. This is a necessary step to obtain a single adherent population of FLS, as freshly collected synovial tissues contain a variety of cells, including lymphocytes and macrophages. The adherent FLS were split at a 1:3 ratio under sub-confluent and passaged conditions. The FLS were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (FUJIFILM Wako Pure Chemical Co., Osaka, Japan) supplemented with 10% fetal bovine serum (FBS) (Gibco, Waltham, MA, USA) and penicillin/streptomycin (FUJIFILM Wako Pure Chemical Co.). The FLS were used for experiments at passages 3–6. For cytokine stimulation using recombinant human TNF-α, IL-1β, IL-6, sIL-6R, and IL-17 (Biolegend, San Diego, CA, USA), the FLS were starved in DMEM with 0.1% FBS for 24 h before the addition of recombinant proteins.
The immortalized FLS from patients with RA (MH7A cells) were obtained from KISSEI Pharmaceutical Co., Ltd. (Matsumoto, Japan). The MH7A cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (FUJIFILM Wako Pure Chemical Co.) supplemented with 10% FBS and penicillin/streptomycin.
RNA extraction and RT-qPCR
Total RNA was extracted and purified from whole synovial tissues and cultured cells using TRIzol reagent (Life Technologies, Carlsbad, CA, USA). For miRNA purification and next-generation sequencing preparation, a Direct-zol RNA kit (Zymo Research, Irvine, CA, USA) with TRIzol reagent was used. For cDNA synthesis from mRNA, PrimeScript RT Reagent Kit with gDNA Eraser (Takara Bio, Kusatsu, JAPAN) was used. RT-qPCR was performed with Brilliant II SYBR Green QPCR Master Mix (Agilent, Santa Clara, CA, USA) using the CFX Connect Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, USA). The primers used in this study are listed in Table 1. The gene expression levels were normalized to the housekeeping gene, glyceraldehyde 3-phosphate dehydrogenase (GAPDH). For cDNA synthesis from miRNA and RT-qPCR, TaqMan MicroRNA Assay kits (Applied Biosystems, Waltham, MA, USA) were used according to the manufacturer’s instructions. The TaqMan probes used are listed in Table 2. The data were normalized to the endogenous reference gene, U6 snRNA.
Next-generation sequencing (small RNA-seq)
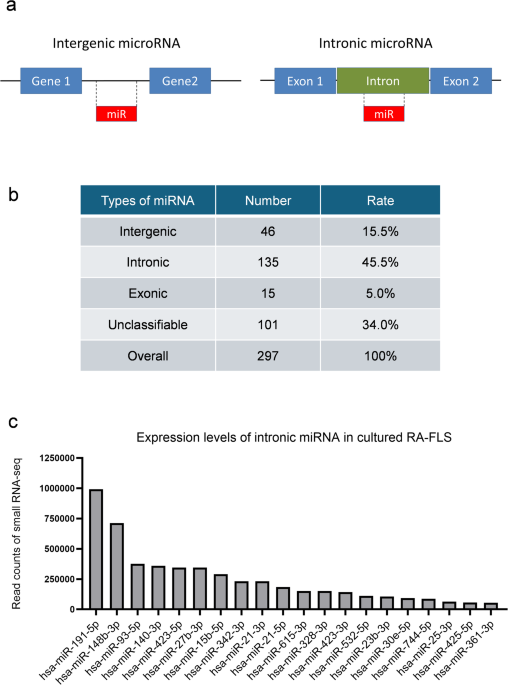
Small RNA sequence library preparation, sequencing, mapping, and gene expression analyses were performed using DNAFORM Inc. (Yokohama, Kanagawa, Japan). The quality and quantity of the extracted miRNAs were assessed using the QuantiFluor RNA System (Promega, Madison, WI, USA) and Agilent Small RNA kit of the BioAnalyzer 2100 System (Agilent Technologies) according to the manufacturer’s instructions. A small RNA-seq library was prepared using the QIAseq miRNA Library Kit (Qiagen, Venlo, Netherlands), following the manufacturer’s instructions. In brief, a pre-adenylated DNA adapter was ligated to the 3’ ends of all miRNAs. An RNA adapter was ligated to the 5’ end of mature miRNAs. Subsequently, reverse transcription (RT) was performed using RT primers containing Unique Molecular Indices. Finally, the library was amplified using the sample-specific index primers. The libraries were sequenced on a NextSeq 500 sequencer (Illumina, San Diego, CA, USA) to generate 75 nt single reads. Duplicate reads were removed from the raw reads using Seqkit (ver. 0.10.1, https://bioinf.shenwei.me/seqkit/), and the adapter sequence was trimmed using Cutadapt (ver. 1.16, https://pypi.org/project/cutadapt/). The trimmed reads were mapped to the human genome (the genome assemblies used were GRCh38.p13) using STAR (ver. 2.7.2b, https://github.com/alexdobin/STAR/releases). The reads mapped to the miRNA genes were quantified using featureCounts in the Subread package (ver. 1.6.1, https://subread.sourceforge.net/). The miRNAs were classified as intergenic, intronic, exonic, or others (unclassifiable) using the UCSC genome browser (human, hg38, https://genome-asia.ucsc.edu/index.html, accessed on April/13/2024).
SiRNA and MiRNA mimic transfection into primary cultured FLS and MH7A cells
The siRNA against STAT3 (FlexiTube siRNA, SI02662898), siRNA against RP2 (FlexiTube siRNA, SI00093205) and siRNA negative control (Allstar Negative Control siRNA) were purchased from Qiagen. The miRNA mimics, hsa-miR-21-5p and hsa-miR-27b-3p, and Synthetic Tough Decoy (S-TuD) against miR-21-5p were purchased from Ajinomoto Bio-Pharma (Osaka, Japan). MiR control (MISSION miRNA negative control) was purchased from Sigma-Aldrich (St. Louis, MO, USA). For these small RNA transfections, primary cultured FLS and MH7A cells were treated using Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. Harvested cells and supernatants were used in subsequent experiments.
Next-generation sequencing (conventional RNA-seq)
Conventional RNA sequence library preparation, sequencing, mapping, and gene expression analyses were performed using DNAFORM Inc. Total RNA quality was assessed using a BioAnalyzer (Agilent Technologies) to ensure that the RNA integrity number was greater than 7.0. Double-stranded cDNA libraries (RNA-seq libraries) were prepared using the SMART-Seq Stranded Kit (Clontech, Kusatsu, JAPAN, Cat. #634442) and DNBSEQ MGIEasy Universal Library Conversion Kit (MGI Tech, Shenzhen, China), according to the manufacturer’s instructions. The RNA-seq libraries were sequenced using paired-end reads (150nt of read1 and read2) on a DNBSEQ-G400RS instrument (MGI Technology). The raw reads obtained were trimmed and quality-filtered using Trim Galore! (version 0.6.7, https://github.com/FelixKrueger/TrimGalore), Trimmomatic (version 0.39, http://www.usadellab.org/cms/?page=trimmomatic), and Cutadapt (version 3.7, https://pypi.org/project/cutadapt/). The trimmed reads were mapped to the human GRCh38 genome using the STAR software (version 2.7.10a, https://github.com/alexdobin/STAR/releases). The reads on the annotated genes were counted using featureCounts (version 2.0.1, https://subread.sourceforge.net/featureCounts.html). The transcripts per million (TPM) and the fragments per kilobase of exon per million reads mapped (FPKM) values were calculated from the mapped reads by normalizing them to the total counts and transcripts. The genes with read counts < 5 were excluded and normalized to those of housekeeping genes. The relative expression values were calculated using the following formula: (1) Log2[(the gene read count of each sample)/(total average count of the gene)] and (2) the difference between the mean value of (1) in miR-21-5p mimic-transfected cells and that in miR control-transfected cells. False discovery rate (FDR) was controlled using the two-stage linear step-up procedure of the Benjamini, Krieger, and Yekutieli method to determine an optimal threshold for statistical significance. The upregulated and downregulated genes were identified based on larger fold-changes.
Western blotting
Before sample preparation, MH7A cells and FLS were washed with PBS. The cells were homogenized in 2% sodium dodecyl sulfate sample buffer using BioMasher II (Nippi Corporation, Tokyo, Japan). Subsequently, the samples were centrifuged at 15,000 × g for 5 min at 20–25 °C, and the supernatant was collected. The lysates were electrophoresed on SuperSepAce 5–12% precast gels (FUJIFILM Wako Pure Chemical Co.) and transferred to polyvinylidene fluoride (PVDF) membranes. The membranes were probed with the following primary antibodies: anti- PDCD4 antibody (sheep polyclonal, R&D systems, Minneapolis, MN, USA), anti-SEMA5A antibody (rabbit polyclonal, Genetex, Irvine, CA, USA), and anti-human β-actin antibody (mouse monoclonal, clone AC-15, Sigma-Aldrich). Subsequently, the cells were treated with horseradish peroxidase (HRP)-conjugated secondary antibodies (host: donkey; Jackson ImmunoResearch, West Grove, PA, USA). The HRP activity was detected using the Image Quant LAS 500 system (Cytiva, Tokyo, Japan) with ECL Prime reagent (Cytiva).
Cytometric bead array assay
The culture supernatants of MH7A cells and FLS treated with either miR-21-5p mimic or miR negative control for 48 h were harvested for sample preparation as described above. The OPG levels were measured using the LEGENDplex system (BioLegend) according to the manufacturer’s protocol.
IHC staining for formalin-fixed paraffin-embedded (FFPE) synovial tissues
The sections were obtained from FFPE synovial tissues. After deparaffinization and antigen retrieval in Tris-EDTA buffer (pH 9.0), the specimens were first incubated with primary antibodies (anti-RP2, rabbit polyclonal, Proteintech (Rosemont, IL, USA), anti-PDCD4, sheep polyclonal, R&D Systems), and then with biotin-conjugated secondary antibodies purchased from Jackson ImmunoResearch. The slides were then treated with alkaline phosphatase (AP)-conjugated streptavidin (Jackson ImmunoResearch) and visualized using ImmPACT Vector Red Alkaline Phosphatase Substrate (Vector Laboratories, Burlingame, CA, USA). The slides were counterstained with hematoxylin solution.
IVT mRNA transfection into MH7A cells
A plasmid encoding HA-tagged full-length PDCD4 (NM_014456.5) or an empty control vector (mock) based on the pcDNA3-A(124) vector was constructed, and in vitro transcribed (IVT) mRNA was prepared as previously described40. The insert fragment of the HA-tagged PDCD4 open reading frame was purchased from GenScript (Piscataway, NJ, USA). The IVT mRNA was transfected into cells using Lipofectamine Messenger MAX (Invitrogen), according to the manufacturer’s instructions. Briefly, IVT mRNA was blended with the Lipofectamine reagent in a ratio of 1:3 (pmol mRNA: µL Lipofectamine) in serum-free RPMI, and subsequently incubated for 20 min at 25 °C for complex formation. After transfection, the cells were harvested for subsequent experiments.
Cell viability assay using calcein-AM
MH7A cells (5.0 × 103 cells/well) were seeded in 96-well plates using 10% FBS supplemented RPMI medium. The cells were cultured in each well for 24 h and then transfected with either miR control, miR-21-5p mimic, mock IVT mRNA, or PDCD4 IVT mRNA, as described above. The cell viability was assessed using calcein-AM (Cell Counting Kit-F; Dojindo Laboratories, Kumamoto, Japan) at 0, 24, 72, and 96 h after transfection, according to the manufacturer’s protocol. Calcein fluorescence was quantified using a SpectraMax iD3 system (Molecular Devices, San Jose, CA, USA) at excitation and emission wavelengths of 485 and 525 nm, respectively.
Statistical analysis
The statistical differences between pairs of groups were assessed using an unpaired t-test (Student’s t-test) or a non-parametric Mann–Whitney U-test. To complement the hypothesis testing with estimation, we also calculated the effect size (Cliff’s delta for Mann–Whitney U-test) and interpreted the values greater than 0.33 as indicating a moderate or larger effect. The differences between three or five groups were determined using the Kruskal–Wallis test, followed by the Dunn test. Data processing and analyses were performed using GraphPad Prism 10 (version 10.4.0, https://www.graphpad.com/; GraphPad Software Inc., La Jolla, CA, USA). Data are presented as the mean ± standard deviation or mean ± standard error of the mean.